Jupyter Environment
Important
The JupyterHub environment is currently under active development. Send your feedback to hpc-support@kifu.hu
Accessing JupyterHub
On the JupyterHub portal, you can run Jupyter notebook containers on the Komondor supercomputer through a web interface using the SLURM scheduler.
Access requires eduID identification and valid HPC portal project registration.
The JupyterHub web interface is available at this link: Jump to JupiterHub
After logging in with the eduID password, there’s a second factor also required. Currently, only email authentication can be selected.
After clicking on the email button, the system sends a code that must be entered in order to log in.

Using JupyterHub
Several notebook servers can be started on the system at the same time from the JupyterHub control panel. Each notebook is a SLURM job that schedules a Singularity container to run. Jupyter servers can be named. If no naming has been written yet, only the default server can be started. You can also view the parameters of active scheduled jobs on the interface below. You can access the running containers by clicking on the access link. You can even save the links to your browser’s bookmarks for convenient return.
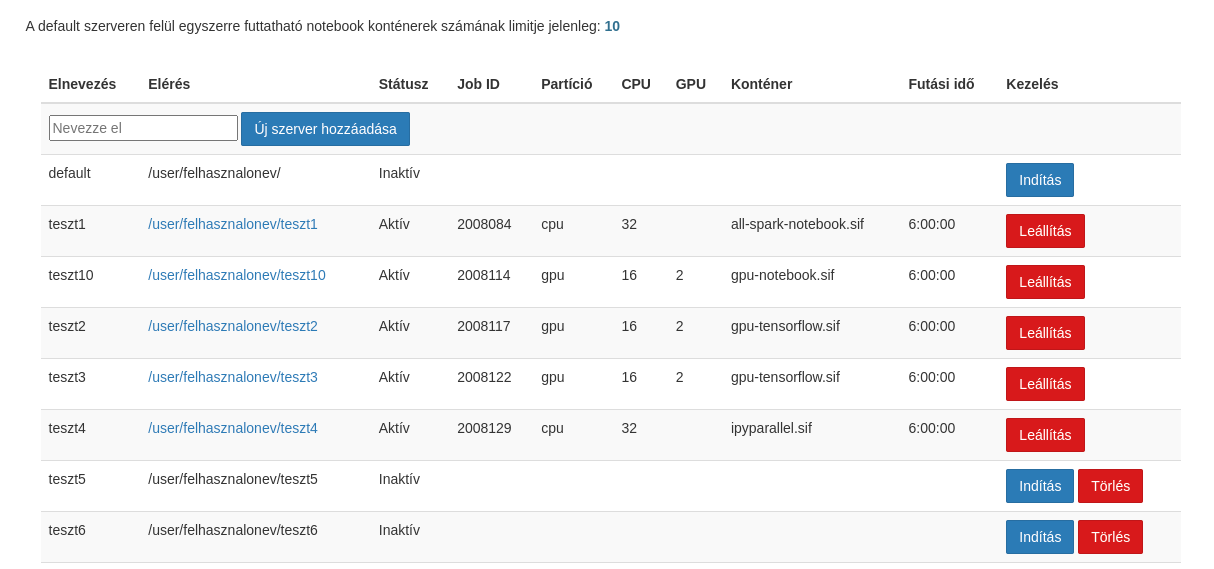
The options can be selected in the next step after pressing the start button. The resource usage is deducted from the quota. The server will run until the time specified at startup, unless it is stopped manually by the user.
Note
It may happen that the physical machines are in a standby state due to energy management concerns. In this case, the startup of the container can take up to 10-15 minutes.
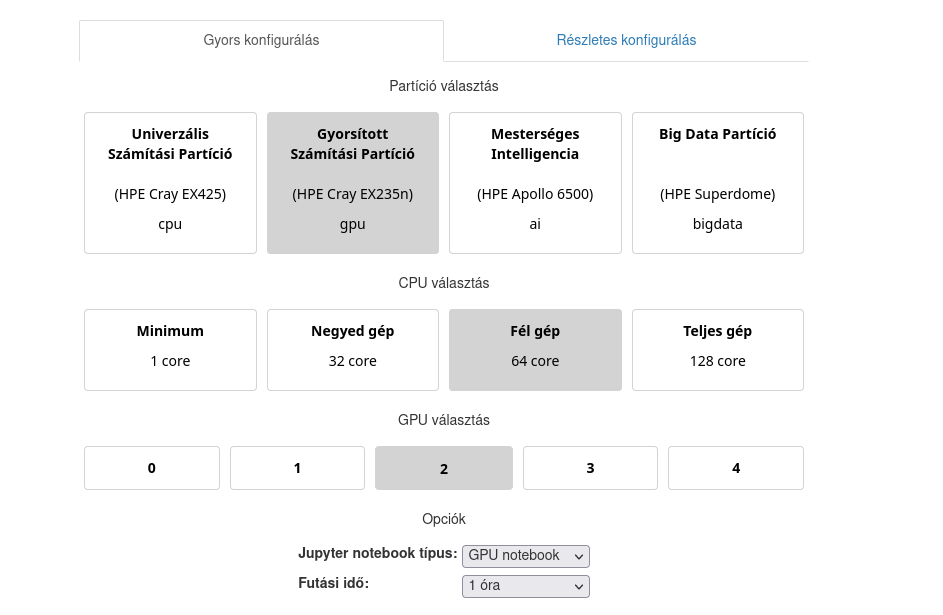
The advanced configuration option is also available, where, for example, the cpu, gpu, and memory settings can be fine-tuned. The job duration can be altered as well.
Hint
If the container stops immediately after starup, it is possible that the job did not get enough memory to use the Jupyter environment. Jump to the memory allocation chapter The job parameters can be corrected in the detailed configuration tab.
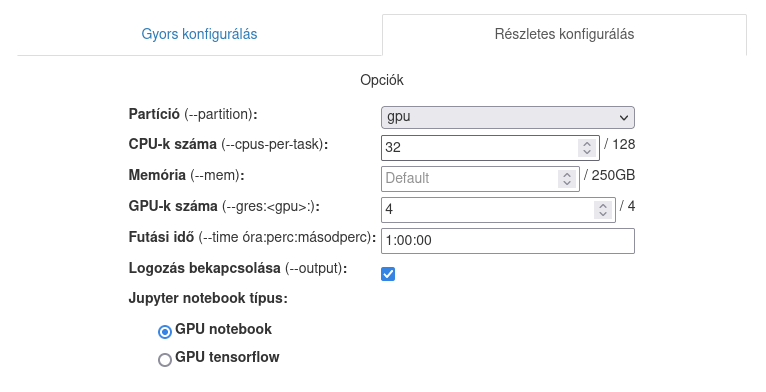
After the notebook started, you can select from the activities, taking notes, editing the code or running a terminal window. You can access the control panel by selecting the Hub Control Panel under the File menu item.
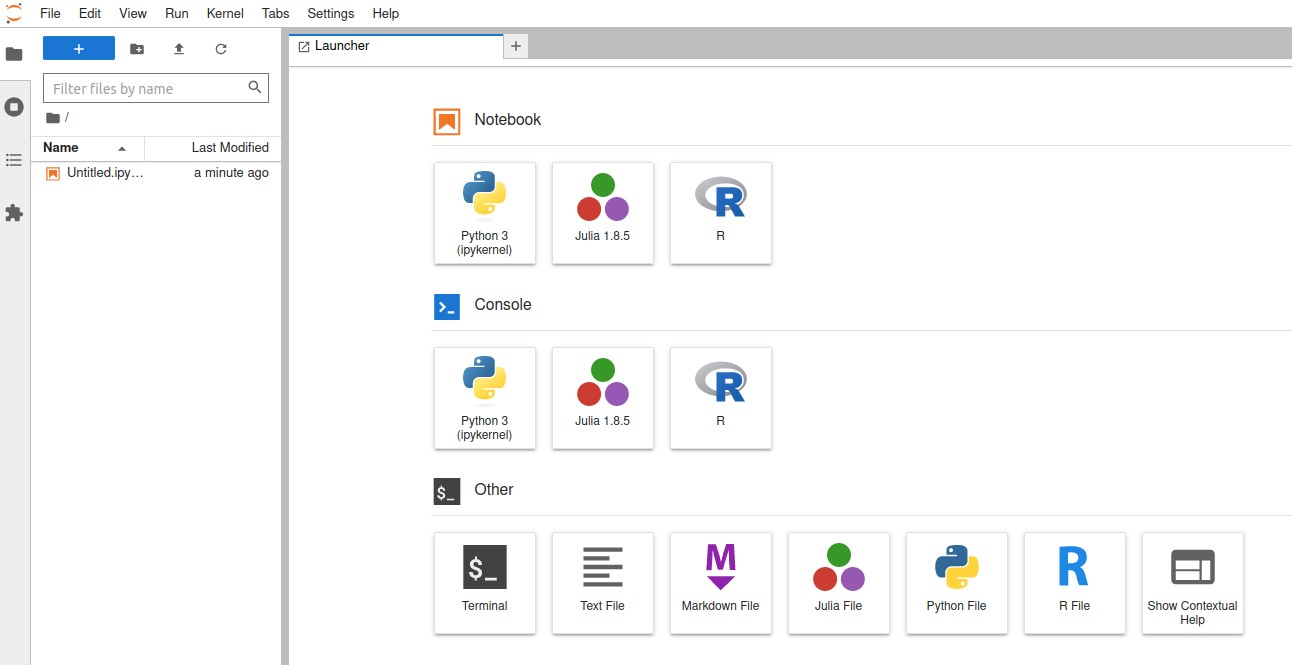
The home directory from the login node is automatically mounted in to the container. The notes and codes saved on the web interface will be found in the $HOME/notebooks directory, and also can be copied here and accessed at any time during the Jupyter sessions.
Jupyter notebook documentation in English can be found here: Go to Jupiter Docs
Stopping the Jupyter container
If jupyter notebook is still running, but you no longer need it, you can stop it manually before the specified time. You can return to the control panel from a running container by selecting Hub Control Panel from the menu.
By clicking on the red Stop button, the slurm job will be canceled immediately. In this way, it can be ensured that a container will no longer consume CPU time.
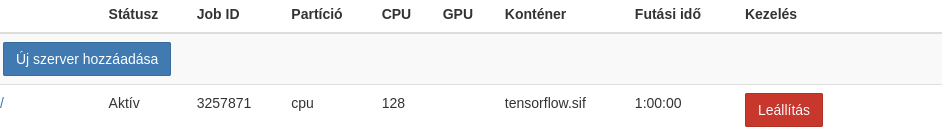
List of available containers
Datascience
Python3 IPykernel (IPython interpreter), IRKernel (R interpreter), IJulia (Julia interpreter)
curl, git, nano-tiny, TeX Live, tzdata, unzip, vi-tiny, wget
altair, beautifulsoup4, bokeh, bottleneck, cloudpickle, openblas, cython, dask, dill, facets, h5py, ipympl, ipywidgets, jupyterlab-git, matplotlib-base, numba, numexpr, openpyxl, pandas, patsy, protobuf, pytables, scikit-image, scikit-learn, scipy, seaborn, sqlalchemy, statsmodel, sympy, widgetsnbextension, xlrd
caret, crayon, devtools, forecast, hexbin, htmltools, htmlwidgets, nycflights13, randomforest, rcurl, rmarkdown, rodbc, rpy2, rsqlite, shiny, tidymodels, tidyverse, unixodbc
HDF5.jl, Pluto.jl
Tensorflow
Python3 IPykernel (IPython interpreter)
Tensorflow
curl, git, nano-tiny, TeX Live, tzdata, unzip, vi-tiny, wget
altair, beautifulsoup4, bokeh, bottleneck, cloudpickle, openblas, cython, dask, dill, facets, h5py, ipympl, ipywidgets, jupyterlab-git, matplotlib-base, numba, numexpr, openpyxl, pandas, patsy, protobuf, pytables, scikit-image, scikit-learn, scipy, seaborn, sqlalchemy, statsmodel, sympy, widgetsnbextension, xlrd
Pytorch
Python3 IPykernel (IPython interpreter)
Pytorch
Tensorflow
curl, git, nano-tiny, TeX Live, tzdata, unzip, vi-tiny, wget
altair, beautifulsoup4, bokeh, bottleneck, cloudpickle, openblas, cython, dask, dill, facets, h5py, ipympl, ipywidgets, jupyterlab-git, matplotlib-base, numba, numexpr, openpyxl, pandas, patsy, protobuf, pytables, scikit-image, scikit-learn, scipy, seaborn, sqlalchemy, statsmodel, sympy, widgetsnbextension, xlrd
Spark
Python3 IPykernel (IPython interpreter), IRKernel (R interpreter)
Apache Spark, PyArrow
ggplot2, grpcio, grocui-status, sparklyr, rcurl
curl, git, nano-tiny, TeX Live, tzdata, unzip, vi-tiny, wget
altair, beautifulsoup4, bokeh, bottleneck, cloudpickle, openblas, cython, dask, dill, facets, h5py, ipympl, ipywidgets, jupyterlab-git, matplotlib-base, numba, numexpr, openpyxl, pandas, patsy, protobuf, pytables, scikit-image, scikit-learn, scipy, seaborn, sqlalchemy, statsmodel, sympy, widgetsnbextension, xlrd
IPython Parallel
Kernel: Python3 IPykernel (IPython interpreter)
Python Parallel engine
curl, git, nano-tiny, TeX Live, tzdata, unzip, vi-tiny, wget
altair, beautifulsoup4, bokeh, bottleneck, cloudpickle, openblas, cython, dask, dill, facets, h5py, ipympl, ipywidgets, jupyterlab-git, matplotlib-base, numba, numexpr, openpyxl, pandas, patsy, protobuf, pytables, scikit-image, scikit-learn, scipy, seaborn, sqlalchemy, statsmodel, sympy, widgetsnbextension, xlrd
Nvidia Tensorflow
TensorFlow, TensorRT, TF-TRT
CUDA, cuBLAS, cuDNN, NCCL
ETL (DALI, RAPIDS)
Training (cuDNN, NCCL)
Nvidia Pytorch
PyTorch, TensorRT, Torch-TensorRT
CUDA, cuBLAS, cuDNN, NCCL
ETL (DALI, RAPIDS)
Training (cuDNN, NCCL)
GPU AI Lab
Python3 IPykernel (IPython interpreter), IRKernel (R interpreter), IJulia (Julia interpreter)
Jupyter AI
TensorFlow, PyTorch
dask, fastai, keras, rapids, xgboost
CUDA, cuBLAS, cuDNN, NCCL, TensorRT
curl, git, nano-tiny, TeX Live, tzdata, unzip, vi-tiny, wget
altair, beautifulsoup4, bokeh, bottleneck, cloudpickle, openblas, cython, dask, dill, facets, h5py, ipympl, ipywidgets, jupyterlab-git, matplotlib-base, numba, numexpr, openpyxl, pandas, patsy, protobuf, pytables, scikit-image, scikit-learn, scipy, seaborn, sqlalchemy, statsmodel, sympy, widgetsnbextension, xlrd
caret, crayon, devtools, forecast, hexbin, htmltools, htmlwidgets, nycflights13, randomforest, rcurl, rmarkdown, rodbc, rpy2, rsqlite, shiny, tidymodels, tidyverse, unixodbc
HDF5.jl, Pluto.jl
Using your own container
Important
Instructions for creating Jupyter containers can be found at the DKF HPC Gitlab portal. Refer to DKF GitLab jupyter repository
The container must be created under the $HOME directory named as jupyter.sif. It can be run from the JupyterHub server by selecting $HOME/jupyter.sif. You will not be able to start any container without jupyterlab and the patched batchspawner package, these must be included in the container produced by the user.
IPython Parallel
The IPython Parallel environment enables parallel execution from a Python program. The program can be developed interactively on JupyterHub, and later can be run separately in a batch job from the login node.
Examples for parallel execution on several compute nodes can be found on the DKF HPC Gitlab portal. Refer to DKF GitLab ipyparallel repository